Avian Influenza as a model airborne pathogen (all virus types)
Disease description
Avian influenza (AI) is a highly contagious viral disease that primarily affects poultry and wild water birds. It poses a global challenge due to its widespread circulation and high mortality rates. AI can be categorized as either low pathogenic (LPAI) or highly pathogenic (HPAI), depending on its impact on avian species and the molecular characteristics of the virus. HPAI, caused by avian influenza A (H5) and A (H7) viruses, is particularly severe, being both highly contagious and often deadly in poultry.
Outbreaks of HPAI among wild birds and poultry have become increasingly common over the past 15 years, with incidents rising significantly. These outbreaks now occur more frequently across all continents, with a higher concentration in Asia, Europe, and the Americas. Since 2020, there has been a notable increase in both H5Nx and H5N1 cases, driven by a variant of the Gs/Gd H5N1 virus (clade 2.3.4.4b), which has caused unprecedented deaths among wild birds and poultry in Africa, Asia, and Europe. By 2022, the virus had spread to the Americas, affecting poultry farms and infecting a growing number of wild and domestic mammals. In that same year, 67 countries across five continents reported H5N1 HPAI outbreaks in poultry and wild birds to WOAH, resulting in the deaths or culling of over 131 million domestic poultry.
MOOD’s case study dedicated to Avian Influenza aims to design and develop sustainable Epidemic Intelligence tools to meet the needs of Public and Veterinary Health practitioners for detection, monitoring, risk assessment and response activities.
Disease profile
To reach a comprehensive understanding of the factors affecting the ecology, distribution, and trend in Europe of the infectious diseases included in MOOD, information from the available literature and expert input was collected and summarized in “disease profiles”. These documents synthesize the available knowledge on specific diseases, such as HPAI (In Press).
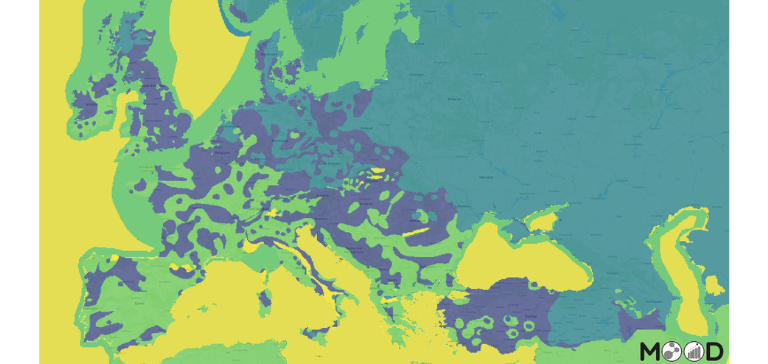
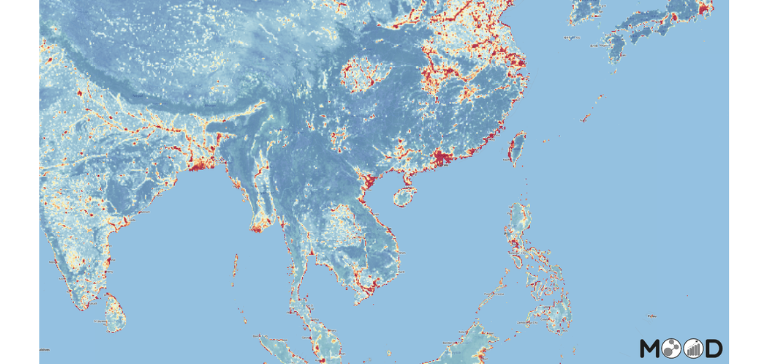
Risk maps
Risk maps and modelling outputs available on MOOD platform under Avian Influenza disease section:
Publications
- Comparing the worldwide distribution of highly pathogenic avian influenza H5 risk from 2020 onward to its previous historical distribution. Dupas MC*, Vincenti-Gonzalez M.F*, Artois J, Dhingra M, Guinat C, Vergne T, Wint W, Hendrickx G, Marsboom C, Gilbert M, Dellicour S. (*) denotes equal contribution.
- Poultry intensification and emergence of highly pathogenic avian influenza. Dupas MC*, Vincenti-Gonzalez MF*, Artois J, Dhingra M, Erazo D, Marsboom C, Dellicour S, Gilbert M (*) denotes equal contribution.
Case study leader

Université Libre Bruxelles, SpELL lab, Maria Fernanda Vincenti Gonzales (maria.fernanda.vincenti.gonzalez@ulb.be)
For more video content about HPAI
In this presentation, Sarah Valentin presents the results of our recently pusblished paper which focuses on how outbreak-related information disseminates from a primary source to to a definitive aggregator, an EBS tool. She and her team analysed news items reporting avian influenza outbreaks in birds worldwide between July 2018 and June 2019 and detected by PADI-web and HealthMap.
Bahdja Boudoua presented her work which’s objective was to understand how health information (signal) is disseminated from a primary source (transmitter) to a final source (EI system) through quantitative and qualitative network analysis methods.